Intraoperative Multisensory Tissue Differentiation in Oncology
Project Details
- Funding Body: DFG
- Date: 2020
- Online: https://www.grk2543.uni-stuttgart.de/en/
Project Story
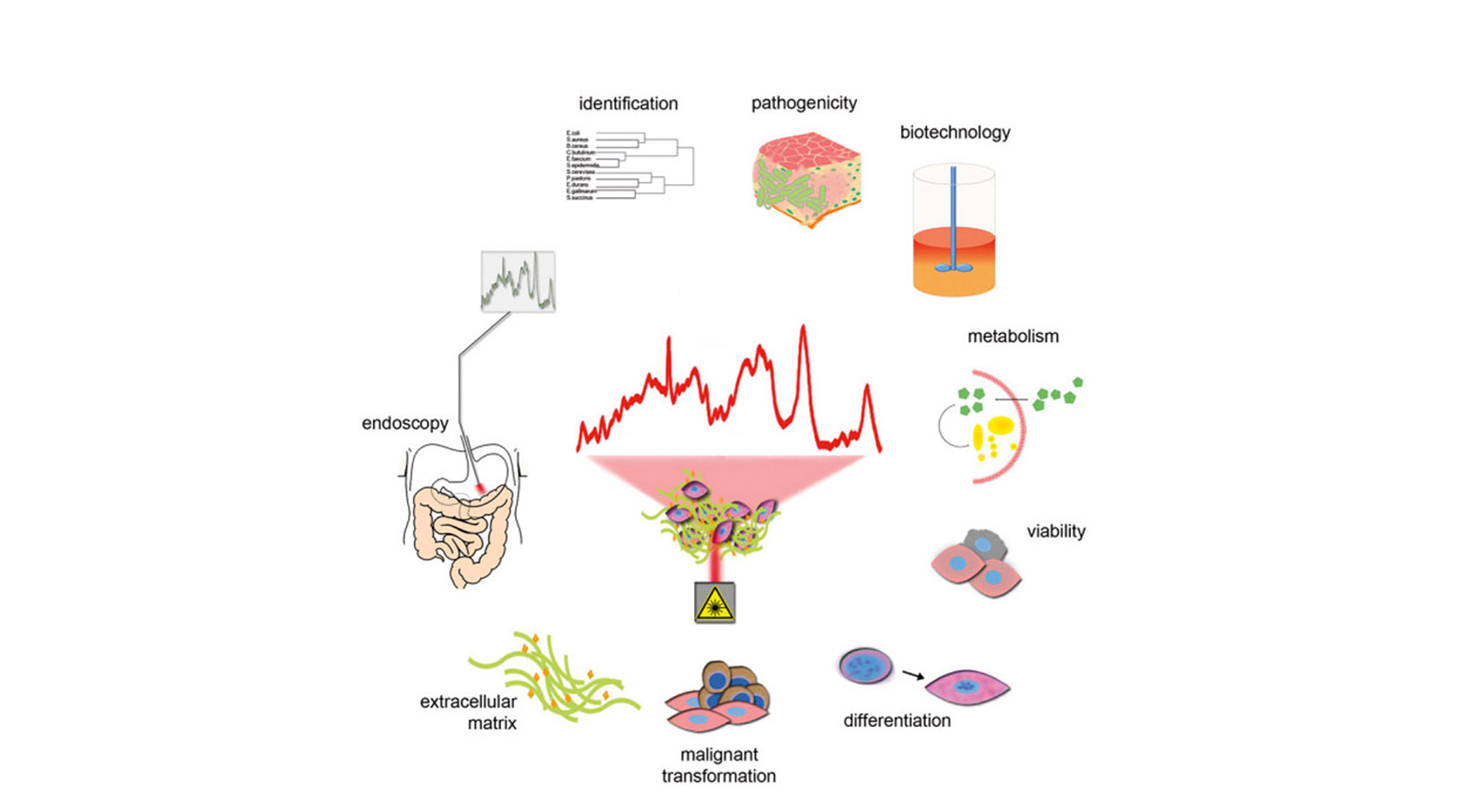
New surgical methods aim to minimize invasiveness, morbidity and duration of treatment while simultaneously maximizing treatment effectiveness. During these surgical interventions, a reliable identification of target structures and surrounding tissue is of utmost importance for achieving this objective, particularly in the field of oncology. Current innovations significantly improve pre- and postoperative diagnostics for distinguishing between benign and malignant tissue structures. Today, refined imaging systems can guide presurgical decision-making more accurately, while enhanced histopathological examination methods allow for a reliable classification of the dissected tissue after surgery. This research training group, however, focuses on intraoperative tissue identification. The fusion of novel multimodal sensor systems by means of machine learning offers a high potential for new procedures to discriminate between tissues that goes beyond the information content of the separate sensor data. The presented methods supplement the gold standard of intraoperative tissue identification, i.e. frozen section diagnostics. As a result, we provide additional information to support surgeons in their decision-making between resection and tissue preservation.
Epigenetic changes often play a role in pathological tissue changes. DNA methylations and histone modifications are dynamic regulators of proliferation and cell behavior and thus have an influence on tissue morphology in the pathogenesis, diagnostics, and prevention/therapy of various oncological diseases. In comparison to healthy cells, tumor cells show an altered epigenetic profile. DNA methylations in malignant tumors are the best studied so far. Tumor cells often show so-called hypomethylations of gene promoter regions, which leads to an activation of the affected genes. The DNA methylation profile can be essential for the pathogenesis, diagnosis and therapy of the individual tumor disease. Novel therapeutic methods, which specifically modulate the dynamic epigenetic network of malignant cells, have great potential to be used in the differentiation of oncological tissue in the future.
New methods for the identification and analysis of cell-specific DNA methylation profiles would contribute to the understanding of the pathogenesis and diagnostics of oncological diseases and may support the development of new therapies. The problem here is that established methods for the investigation of epigenetic alterations have so far been very time-consuming and require the disruption of the tissue and the isolation of the cellular DNA.
Raman spectroscopy is a non-invasive analytical technique that allows molecular changes to be studied at the single cell level. Laser light is used here to excite specific molecular vibrations, which allows the specific Raman scattering to be detected. Chemical and structural changes can thus be tracked in situ and in real time at high resolution (down to about 200 nm) using Raman spectroscopy. Our own preliminary work indicates the possibility of non-invasive and marker-free analysis of epigenetic changes, such as differentiation between methylated and unmethylated DNA regions. To date, changes in nucleic acid structures have not been investigated in situ in cell cultures or tissues using Raman microspectroscopy.